

Understanding and engineering the logic behind plant decisions
This research is proudly funded by the Netherlands Organisation for Scientific Research (NWO) Open Technology Programme, fostering innovation in plant biotechnology.
Interviews and features about our research
Aschwin Tenfelde interviews us about the project
Read the article
Interview on Humberto, AVROTROS, 19 September 2025. Discussing the NWO-funded research on flower pigmentation patterns.
Listen to the Interview on NPO Radio 1Elisabetta Santangelo from AD interviews us about the project
Read the article
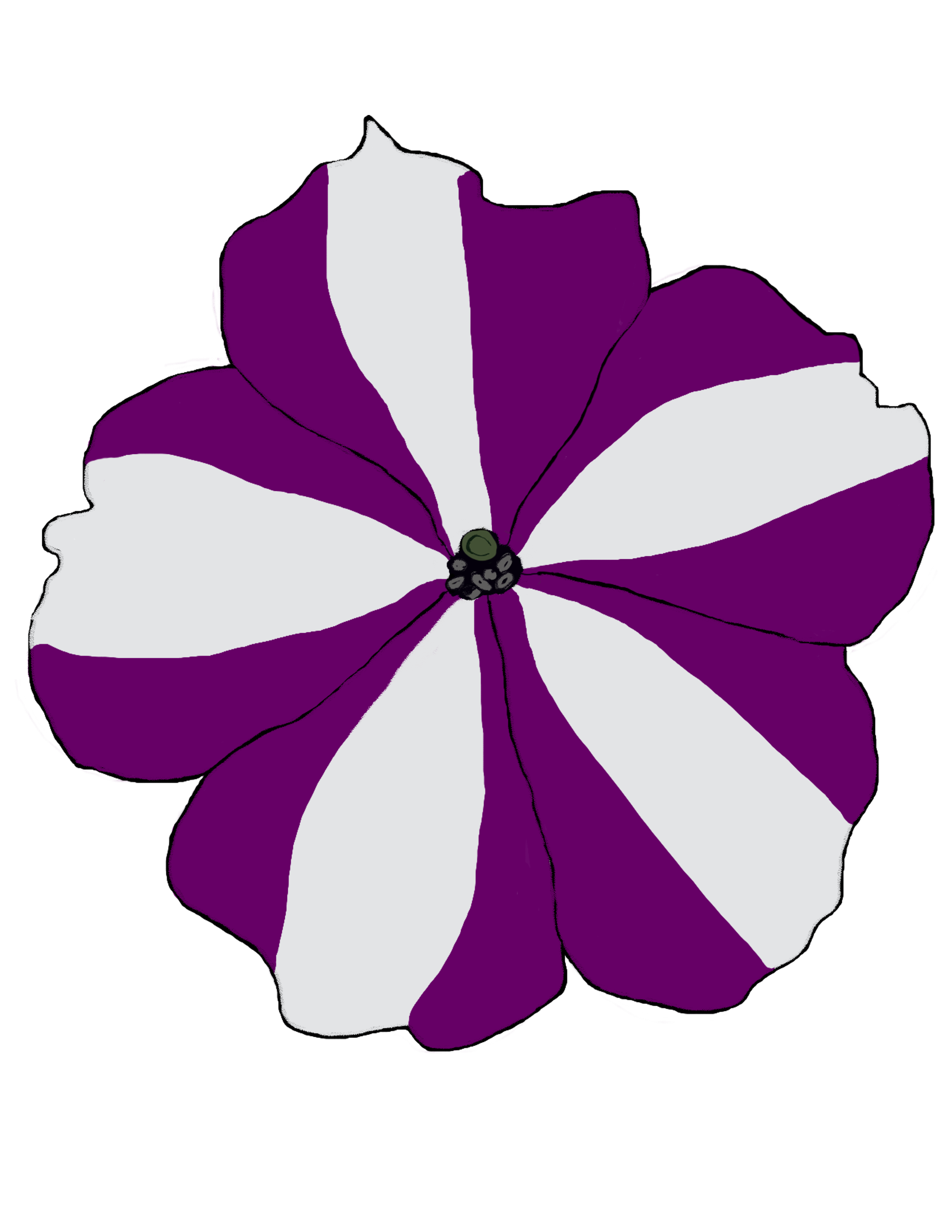
Understanding how sRNAs and epigenetic marks create and maintain complex patterns of cell identity in flower pigmentation
Modern plant breeding relies heavily on identifying new traits from breeding populations that can be associated with genetic (DNA sequence) markers. However, some traits are driven by changes in sRNAs or epigenetic marks, making them harder to associate with a specific DNA sequence alteration. Thus, maintaining such traits is a significant challenge. Our project aims to use flower pigmentation as a visual marker in combination with genetic screens, advanced omics techniques, visualisation of sRNA movement, and genome editing to understand and exploit mechanisms regulating variation driven by sRNAs and epigenetic marks. This will open new opportunities for introducing novel traits and speeding up crop improvements, ultimately leading to economic impact for breeders and farmers.
Multicellular organisms develop from clusters of identical cells with the same genome. These cells can differentiate in gene expression due to internal or external cues, using mechanisms such as epigenetic marks, transcription factors, and small RNAs (sRNAs). While some factors involved in generating these patterns have been identified, we still lack a comprehensive understanding of how these patterns are induced in response to developmental or environmental cues and how distinct gene expression patterns are established and maintained. This knowledge gap hinders our ability to leverage a plant's natural adaptability to environmental cues and develop new traits for breeding programs.
This proposal aims to study flower pigmentation patterns, especially in ornamental plants like petunias, chrysanthemums, and orchids, as a model for understanding the development and maintenance of complex patterns of cell identity. In specific petunia cultivars, patterns of pigmented and unpigmented cells are driven by Post-Transcriptional Gene Silencing (PTGS) of CHALCONE SYNTHASE (CHS), a key enzyme in the synthesis of the anthocyanin pigment, through sRNA-mediated RNA interference (RNAi). Using genetic screens, advanced omics techniques, visualisation of sRNA movement, and genome editing we will explore how stable sRNA populations are formed and maintained to apply this knowledge to improve breeding strategies across both ornamental and non-ornamental crops.
What are the key genetic factors for initiating CHS-silencing in developing flowers?

WP1 Lead

Genetic basis of pattern formation

Coming Soon 2026

Genetic markers and pattern initiation

How is the spatial pattern of CHS silencing maintained independently from its initiation?

WP2 Lead
sRNA dynamics and epigenetic regulation

Coming Soon 2029

sRNA movement and DNA methylation

Industry partners driving innovation in plant breeding

Leading breeder and propagator of Anthuriums, Orchids and Bromeliads with 85 years of experience in creating innovative, sustainable and colorful flowers and plants

Biotechnology company specializing in plant breeding innovations and genetic technologies for agricultural and horticultural applications.

Chrysanthemum breeder developing innovative varieties with unique colors, forms, and improved cultivation characteristics for global markets.

Flower breeding company focused on developing new varieties of chrysanthemum flowers with enhanced aesthetic and commercial value.

Global leader in berry genetics and breeding, pioneering advanced breeding techniques and sustainable growing practices for premium berry varieties.

Global vegetable breeding company developing innovative varieties through advanced breeding techniques and biotechnology for sustainable agriculture.
